- Welcome
- Introduction
- Publications and presentations
- Signing in
- System requirements
-
How To
- Logging into GNomEx
- Submitting an experiment request
- Submitting a microarray hybridization request
- Submitting a sample quality request
- Submitting a sequencing request
- Requesting that microarray hybridization or sequencing be performed on samples previously submitted to sample quality testing
- Adding lanes to a previously submitted sequencing request
- Registering an external experiment in GNomEx
- Finding and viewing a summary of an experiment
- Editing an experiment
- Downloading experiment results
- Linking the samples from an experiment to their BST sample
- Viewing analyses
- Downloading analyzed data files
- Viewing a description of the materials and the methods used during processing of a specific kind of experiment
- Performing a search for experiments and analyses meeting a specific type of criteria
- Submitting a work authorization
- How pricing and billing works in GNomEx
- Adding a new price criterion
- Creating a new price sheet
- Creating a new price category
- Modifying an existing price sheet
- Modifying an existing price category
- Modifying an existing charge item
- Preparing and sending out invoices for clients
- Generating general ledger interface data for automatically billing university accounts
- Window, Tab, and Field Descriptions
-
GNomEx Error Messages and Information Windows
- Are you sure you want to delete all the hybridizations?
- Are you sure you want to delete all of the samples?
- At least one non-empty file must be selected in order for download to proceed.
- Please enter all of the required fields for the hybridization.
- Please enter all required fields for the samples.
- Please enter at least one search criterion.
- Please select a price category.
- Please select a price sheet.
- Remove all hybs currently showing in the list?
- Remove all samples currently showing in the list?
- The number of samples to setup has changed. Do you want to clear out the existing samples?
- The request has not been saved. Are you sure you want to quit?
- The request is now assigned to project.
- The webpage you are viewing is trying to close the window.
- Unable to generate GL interface. Cannot find approved folder.
- You are no longer logged on. Please logon again.
- Your changes have not been saved. Are you sure you want to exit?
-
Working With the List of Samples Submitting for Microarray Hybridization
- Adding a Sample to the List of Samples You Will Be Submitting for Microarray Hybridization
- Adding a sample to the list of samples you will be submitting for microarray hybridization that is similar to another sample already in the list
- Deleting a sample from the list of samples you will be submitting for microarray hybridization
- Deleting all of the samples currently included in the list of samples you will be submitting for microarray hybridization
-
Working With the List of Hybridizations By Requests
- Adding a hybridization to the list of hybridizations you will be requesting
- Adding a hybridization to the list of hybridizations you will be requesting that is similar to another hybridization already in the list
- Deleting a hybridization from the list of hybridizations you will be requesting
- Deleting all of the hybridizations currently included in the list of hybridizations you will be requesting
- Editing a hybridization from the list of hybridizations you will be requesting
- Working with the list of hybridizations you will be requesting
- The hybs tab (submit microarray hybridization request window)
- The submit microarray hybridization request window
- Submitting a microarray hybridization request
- Microarray hybridization requests
-
Specialized Role Topics
- Uploading a sample sheet
- Setting/changing the prices of a charge item
- Setting/changing the experiment type(s) associated with a price sheet
- Setting/changing the criteria by which a charge item will be applied to billing for an experiment
- The sample view tab (new experiment module)
- The hyb setup tab (new experiment request module)
- The hyb view tab (new experiment request module)
- The new project window
- The edit project window
- Adding a new project while you are entering a microarray hybridization request
- Editing an existing project while entering a microarray hybridization request
- Adding a new sample characteristic for annotation while submitting a sample hybridization
- Editing an existing sample characteristic for annotation while submitting a sample hybridization
-
Guides
- User Guide
- Experiment orders
- The foundation of GNomEx
- Experiments at a glance
- Submitting a HiSeq or MiSeq experiment order
- ABI Capillary (Sanger) Sequencing
- Checking on experiment progress
- Copy and pasting into samples grid
- Posting outside experiment data
- iScan experiments
- Downloading and uploading your data
- The download files window
- Downloading large data sets
- Downloading chromatograms
- Fast data transfer from the command line
- Troubleshooting guide for downloading
- Launching FDT with Webstart
- Uploading your data
- Annotating your experiment
- Associating experiment files back to the samples
- Sample annotations
- Copy and paste for the samples grid
- Bulk sample sheet import
- Analysis and data tracks
- Create an analysis
- Data tracks
- Disk usage
- Configuring GNomEx
- Context-sensitive help
- Customize the billing account fields
- Dictionaries
- Seq lib protocols and barcode schemes
- Configuring HiSeq rapid mode sequencing options
- Developer Guide
- Installation Guide
- Configuring GNomEx for FDT
- Configuring GNomEx to use LDAP Authentication
- Compiling and Building
- The Database Schema
- GNomEx Open Source
- Developer Documents
- Illumina HiSeq Realtime Info — The Metrix Server
- FAQs
- Demo
GNomEx now serves as a repository for files visualized in the following genome browsers:
- IGB
- IGV
- UCSC Genome Browser.
GNomEx Raw experiment files -> GNomEx Analysis files -> GNomEx Data Track files -> Genome Browsers
In order to create Data Tracks to distribute for visualization, an Analysis must first be created. Then the specific files supported by the browser (e.g., bam, useq, etc.) can then be dragged to the Data Track file.
How to Navigate to a Data Track
The Data Tracks view is organized by Organism and Genome Build. The view supports a nested folder structure so that like data tracks can be grouped together. You can look up a Data Track by data track number. The data tracks have the number 'DTxxx'.
Properties are ported over to the GNomEx model, so you will be able to define properties using 'Configure Properties' from the GNomEx home page.
How to Create a Data Track
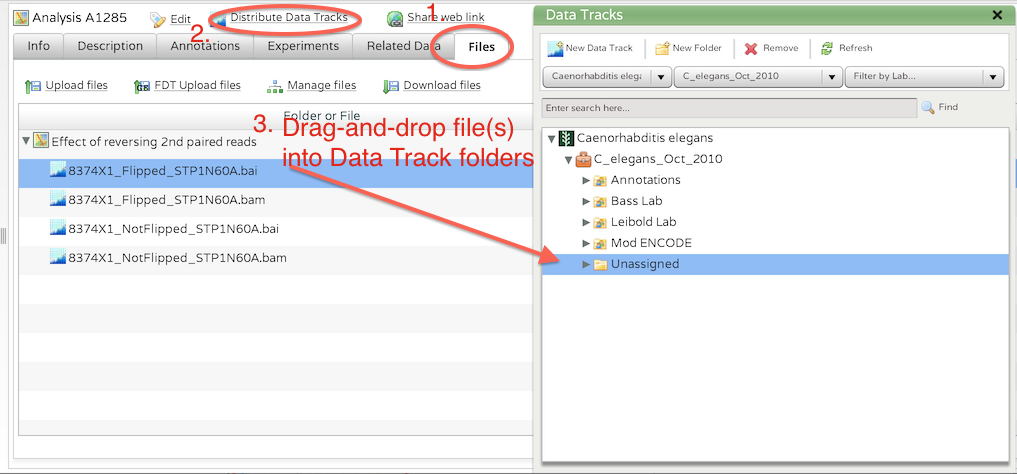
To perform the link, create or click on an existing Analysis and go to the files tab. Click on the 'Distribute Data Tracks' link.
The Distribute Data Tracks window will appear and filter based on the Organism, Genome Build, and Lab group. You may need to deselect the lab group from the dropdown if you want to see all data tracks for this genome build.
Select an analysis file and drag-and-drop it to the data track tree. If you drag it onto an existing data track, it will link the analysis file to that data track. If you drag it over a folder or genome build folder, it will create a new data track, giving it the name of the analysis file.
The Data Tracks window will continue to stay visible after you have linked/created a data track. You can click on another analysis file or another analysis and perform the same operation again.
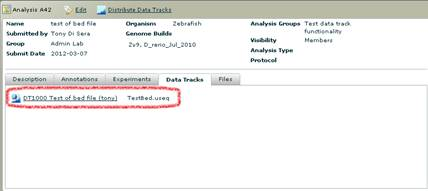
The Analysis Detail view has a Data Tracks tab that will show you links to all of the data tracks. Click on the link to go to the Data Tracks view.
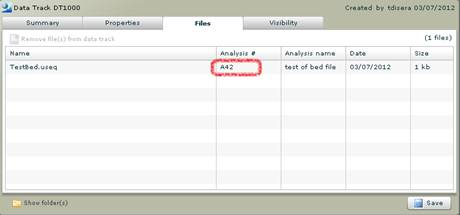
On the Files tab of the Data Track Detail view, the analysis number that the data track file is link appears in the files grid.
When you remove an Analysis or an Analysis file that is linked to data track(s) you will get a message that the data tracks must be unlinked or removed first. Let me know if you would like to happen automatically.
Remember to click the 'Refresh DAS/2 server' when you are ready to distribute the data tracks to the DAS/2 server.
How to visualize the Data Track in a Genome Browser
Data Tracks can be visualized in three genome browsers: IGB, IGV, and the UCSC Genome Browser. To launch IGV or the UCSC Genome Browser, click on the link in the top right corner. (The link will be disabled if the file type is not supported by the browser.)

To launch IGB, click on one of the links that will launch Webstart for IGB. Now you are ready to connect to this data source in IGB. http://myserver.somewhere.edu:8080/das2gnomex/genome. Set up a data source with this URL. The server you want may already be preconfigured. Look for the URL in the available servers and select it if present.

To load DataTracks from within IGV, first create an IGV server within GNomEx by clicking the IGV Browser button after selecting any DataTrack. Copy the URL in the popup.
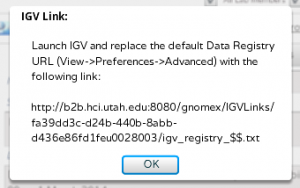
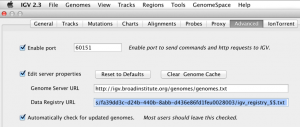
Launch IGV and replace the Data Registry URL with this link (View -> Preferences -> Advanced).
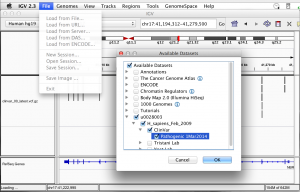
Select tracks to load from within IGV (File -> Load from Server -> click checkbox to expand last server)
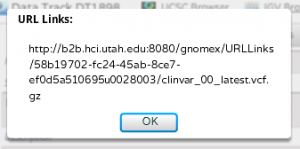
If you wish, use the URL Links button to fetch a URL to the file. This can be pasted into a variety of applications that can Load From URL (IGB, IGV, GBrowse, UCSC).
Lastly, clicking the UCSC Browser button after selecting a DataTrack will open a new window and load the UCSC Browser and your DataTrack as custom tracks. Repeat to add additional tracks. Which UCSC Browser is loaded can be configured in the MyAccount page.




